Highlight
Live Cell analysis of Chromosome Movement and Conformation in Budding Yeast
Achievement/Results
Learning how chromosomes are arranged in the nucleus and how mobile they are in a crowded environment is essential in understanding how chromosomes interact during repair of chromosome damage and in other aspects of chromosome biology. To accomplish this has required an interdisciplinary approach, combining genetics, molecular biology and biophysics to create and analyze the movement of fluorescently tagged spots located at precise positions on a budding yeast chromosome.
To study chromosome dynamics, Brandeis University NSF-IGERT trainee Susannah Messer-Gordon, mentored both by Jim Haber (Biologym, Brandeis) and Jané Kondev (Physics, Brandeis), use an ultra-fast, ultra-sensitive novel microscope (OMX) in John Sedat’s lab at UCSF. The goal is to define chromosome dynamics for a budding yeast chromosome arm, by marking the centromere of the chromosome (indirectly with a closely bound spindle pole body) with a red fluorescent protein (Spc29-RFP) and a site near the HML locus, 90 kb away, on the left chromosome arm, with LacI::GFP tethered to an array of LacO sequences. The motion of the GFP and RFP spots measured at these very short intervals is subdiffusive (on a log-log plot of Means Squared Displacement versus time, the slope is not 1 (indicative of free diffusion) but 0.5, indicative of subdiffusion of spots on a polymer (see Graphics).
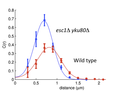
Using this approach the investigators have examined the movements of the left arm of chromosome III and analyzed the effects of removing proteins that normally tether the chromosome end (telomere) to the nuclear envelope. As shown in the Graphic 1, the investigators locate a LacI-GFP (green fluorescent protein) to a site near the left end of the chromosome by its binding to an array of 250 LacO sequences. Similarly they locate the Spc29-RFP (red fluorescent protein) to the spindle pole body that attaches to the chromosome’s kinetochore. A histogram of the distances between the GFP and RFP spots in wild type cells are compared to those lacking the Yku80 and Esc1 proteins that are required to tether telomeres to the nuclear envelope (Graphic 2). The loss of tethering causes a reduction in the mean distance and also a narrowing of the variance, which can be interpreted in terms of polymer models with and without a tether anchoring the end of the chromosome.
Address Goals
This work provides fundamental new insights into how chromosomes in living cells are repaired subsequent to damage. This has important implications for understanding how environmental factors that cause chromosomal damage are repaired, and has implications for cancer biology and evolution. This work required the use of new high microscopes.