Highlight
Challenge Project: Quantification of glycan abundance from mass spec data
Achievement/Results
How is it that prostate and breast cancers metastasize to the bones, while colon cancers migrate to the liver? The answer may have something to do with the way that living cells recognize one another in our bodies. The keys to this cell-cell recognition are elaborate carbohydrate structures that each cell displays on its surface. Migrating cells are able to interpret these marks to determine whether or not they have reached their intended target.
Understanding how cells utilize carbohydrates for cellular recognition has the potential to inform many important areas of biology including immune response, wound healing, vascular disease, tumor formation and metastasis. One of the first steps toward achieving this goal is to quantify the type and number of carbohydrates present in a given type of tissue. We could, for instance, learn that a particular type of tumor is identified with a specific set of carbohydrates. This may, in turn, lead to the development of therapies that would specifically target tumor cells.
Working with Dr. Joseph Zaia at Boston University School of Medicine, Department of Biochemistry, first year IGERT trainees Christopher Jacobs and Jonathan Dreyfuss, and first year NIH Graduate Partnership Program trainee Yevgeniy Gindin developed software specifically tailored to the task of quantifying carbohydrate compositions isolated from biological samples. This work was done in the context of the Boston University Bioinformatics graduate program Challenge Project, a team project class, now in its second year, which was developed as part of an IGERT supported curriculum revision. Projects involve open-ended research using high-throughput data obtained from one of BU’s biology or medical school labs.
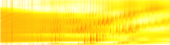
Carbohydrates and other cellular molecules can be analyzed by a technique called mass spectrometry which measures fragments of molecules based on their mass and electrical charge (Fig. 1). Many software packages are available which will analyze mass spectrometry data for proteins, another class of cellular molecules. These software packages work in “discovery mode,” that is, they search through the entire dataset for “mountains” which represent high abundance proteins that stand above the noise. However, the techniques that work for proteins do not carry over well to carbohydrates which are completely different types of molecules. Fortunately, for the research conducted in Professor Zaia’s lab, “which” carbohydrates are of interest is already known. It is the “how much” or abundance of each type that needs to be determined. Such measurements can reveal, for example, that one tissue type has more or less of a critical carbohydrate than another tissue type.
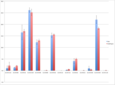
Because existing software is not designed to process mass spectrometry data for carbohydrate abundances, graduate students spend weeks(!) doing the work manually. Jacobs, Dreyfuss, and Gindin, with no previous experience in this field, developed a targeted approach to the problem: they use a list of carbohydate targets to define the areas of interest, the “mountains” (Fig 2.), within the raw data. Then their software calculates and reports the abundance of each carbohydrate by measuring the height and width of its mountain. The targeted approach has the added benefit that it facilitates quick, accurate comparisons of relative abundance between targets and across datasets, an extremely common research practice.
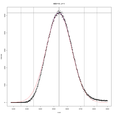
In hopes of making their new tool available to the wider research community, Jacobs, Dreyfuss, and Gindin have developed a free, open source, easy-to-install, and easy-to-use software package that facilitates targeted glycomics analyses (glycomics is the technical name for the study of cell surface carbohydrates). The software has been successfully used to replicate the results (Fig. 3) of previous analyses with improved accuracy and much improved time span, condensing the formerly weeks-long analyses into mere minutes. It also integrates well into current workflows, using only open-source software and formats standardized for the field. A paper describing the software will be featured in an upcoming issue of Analytical and Bioanalytical Chemistry.
Address Goals
This activity primarily addresses the goal of cultivating an outstanding scientific workforce. It emphasizes the goals of creativity, independence, and quality in student research and it creates opportunities for collaborative research among faculty from different disciplines. With respect to the secondary goal of advancing the frontier of knowledge, the work started by the students in this Challenge Project will be carried forward and ultimately advance our knowledge of the role of glycans in cell-cell communication.