Highlight
Arabidopsis meristem/primordia growth analysis framework and meristem 3D reconstruction system
Achievement/Results
The NSF-ChemGen Integrative Graduate Education and Research Trainee Program at University of California, Riverside (ChemGen IGERT) supported research that has led to the development of an Arabidopsis meristem/primordia growth analysis framework and meristem 3D reconstruction system. This was achieved by ChemGen IGERT trainee, Moses Tataw, working with plant developmental biologists Dr. Reddy and electrical engineer Dr. Roy-Chowdhurry, both NSF-funded researchers. Moses’ training is in computer sciences. The Shoot Apical Meristem (SAM) of a plant is made of stem cells that are responsible for the formation of all above ground plant structures. Differentiating cells in the development of SAM form primordia. Primordia develop to become various individual organs. The Reddy lab focuses on studying the dynamics of SAM development and maintenance. In this regard, the group strives to quantitatively represent the relationship between gene expression patterns, cell-division patterns, cell-cell communications, and overall (global) SAM growth.
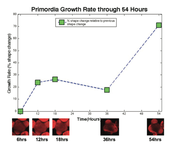
In his project, Moses Tataw tackled the quantitative analysis of SAM growth, with an emphasis on primordial growth patterns. First, Moses developed a method for performing quantitative analysis of primordia development in the model plant Arabidopsis thaliana. This involved learning to grow plants and visualize their meristem with a confocal laser microscope. A contour based approach was used for primordial detection, and a Dynamic Time Warping (DTW) Algorithm was applied to compute the rate of growth. The analysis measured growth in terms of shape change of the primordia stack. The results were published at the International Conference on Image Processing (ICIP) 2010. An example primordia growth curve is shown below. The second phase of this project involves the three-dimensional (3D) reconstruction of entire meristem or individual primordia from two-dimensional image slices obtained with the confocal microscope. Such reconstruction provides the ability to measure primordial or meristem growth based on features like surface area or volume. Such measurements could not be done reliably from the growth computation done in phase one. The 3D reconstruction of an entire meristem and primordia is a significant addition. The figure here shows a reconstructed meristem, with boundaries of 2D slices used for reconstruction that are superimposed on the 3D surface for comparison.
Address Goals
This is a inter-disciplinary research program in identification, spatio-temporal modeling and recognition of dynamical patterns inherent inshoot apical meristem development through the use of novel computational tools in image analysis, statistical modeling, pattern recognition, machine learning and dynamical system identification (From Data to Knowledge). This will lead to the development of new methods for 3D tracking of cells, identifying cell lineages and learning functional models of the dynamics of cell growth and division. The computational tool-kit will enable researchers to gain new insights into the active interplay between cell growth, cell division and changes in gene expression patterns in dynamic developmental fields. This research plan involves a continuous feedback between the biological and computational needs, whereby the developed algorithms evolve based on their interpretation, analysis and validation by the biologists.