Highlight
Protein Nanopatterns by Oxime Bond Formation
Achievement/Results
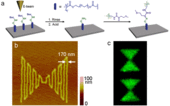
With support by the National Science Foundation, a team of researchers in the Department of Chemistry and Biochemistry at the University of California-Los Angeles, in collaboration with researchers from the Department of Mechanical and Aerospace Engineering at the University of California-Los Angeles, have successfully developed a method to fabricate site-specifically immobilized protein nanopatterns using oxime bond formation. Protein arrays have numerous applications in medicine, ranging from diagnostic arrays to cell and tissue engineering. However, most strategies for immobilizing proteins lead to reduction in bioactivity. As features get smaller, such losses could become increasingly significant. This technique allows for the placement of features with nanoscale resolution and any shape, and may be employed to site-specifically anchor proteins that retain their bioactivity at the nanoscale. This is exceedingly important for production of emerging bioarrays, as feature sizes decrease and fewer proteins are immobilized on each pattern.
The research team synthesized and characterized a polymer containing aminooxy functional groups. This polymer was then patterned on silicon wafers through electron-beam (e-beam) lithography and confirmed nanoscale patterning using atomic force microscopy. The team has shown that proteins modified to contain a reactive ketone at the N-terminus bind to the patterned features with high specificity and in a site-selective manner. This modification can be applied to a wide variety of proteins with maintenance of bioactivity. The ability to immobilize proteins in defined orientations is important because it increases the number of immobilized proteins that remain active on protein nanoarrays. While many techniques exist for fabricating protein nanoarrays, all methods result in either non-specific attachment or are not suitable for long-term applications. The newly developed technique could allow the scientific community to prepare protein nanoarrays with greater sensitivity and lower detection limits than previous immobilization methods.
This work was possible through the funding and opportunities provided by the Materials Creation Training Program (MCTP) IGERT at UCLA. The work required an interdisciplinary team comprised of multiple research groups at UCLA, Prof. Heather D. Maynard and Prof. Yong Chen. Both of these research groups brought different talents and skills. It required the radical polymerization, surface chemistry, and biochemistry expertise of Dr. Maynard, and the expertise of Dr. Chen in patterning polymeric materials using electron-beam lithography. The MCTP IGERT grant provided financial support that allowed a UCLA graduate student, Christopher Kolodziej, to contribute to this project. Without the assistance of the NSF, this work would not have succeeded.
Reference
Christman, K. L.; Broyer, R. M.; Schopf, E.; Kolodziej, C. M.; Chen, Y.; Maynard, H. D.,
Protein Nanopatterns by Oxime Bond Formation, Langmuir, 2011, 27, 1415-1418.
Address Goals
This work offers a significant improvement in protein immobilization methodology, which is critical for advancing protein nanoarray technology, an important area of both diagnostics and tissue engineering.